As suggested by the name Hierarchical Data Format, an HDF5 file is hierarchically structured. The HDF5 group and link objects implement this hierarchy.
In the simple and most common case, the file structure is a tree structure; in the general case, the file structure may be a directed graph with a designated entry point. The tree structure is very similar to the file system structures employed on UNIX systems, directories and files, and on Apple Macintosh and Microsoft Windows systems, folders and files. HDF5 groups are analogous to the directories and folders; HDF5 datasets are analogous to the files.
The one very important difference between the HDF5 file structure and the above-mentioned file system analogs is that HDF5 groups are linked as a directed graph, allowing circular references; the file systems are strictly hierarchical, allowing no circular references. The figures below illustrate the range of possibilities.
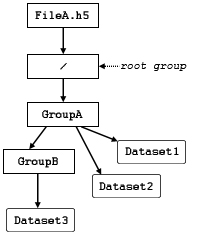
In Figure 1, the group structure is strictly hierarchical, identical to the file system analogs.
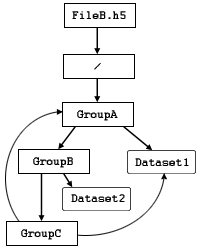
In Figures 2 and 3, the structure takes advantage of the
directed graph's allowance of circular references.
In Figure 2, GroupA is not only
a member of the root group, /,
but a member of GroupC.
Since Group C is a member of Group B
and Group B is a member of Group A,
Dataset1 can be accessed by means of the circular reference
/Group A/Group B/Group C/Group A/Dataset1.
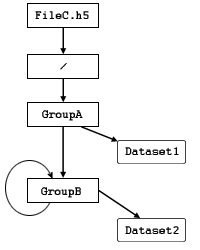
Figure 3 illustrates an extreme case in which
GroupB is a member of itself, enabling a
reference to a member dataset such as
/Group A/Group B/Group B/Group B/Dataset2.

|

|

| ||
| Figure 1: An HDF5 file with a strictly hierarchical group structure | Figure 2: An HDF5 file with a directed graph group structure, including a circular reference | Figure 3: An HDF5 file with a directed graph group structure and one group as a member of itself |
As becomes apparent upon reflection, directed graph structures can become quite complex; caution is advised!
The balance of this chapter discusses the following topics:
Abstractly, an HDF5 group contains zero or more objects and every object must be a member of at least one group. The root group, the sole exception, may not belong to any group.
|
|
| Figure 4: Abstract model of the HDF5 group object |
Group membership is actually implemented via link objects (see Figure 4). A link object is owned by a group and points to a named object. Each link has a name, and each link points to exactly one object. Each named object has at least one and possibly many links to it.
There are three classes of named objects: group, dataset, and named datatype (see Figure 5). Each of these objects is the member of at least one group, which means there is at least one link to it.
|
|
| Figure 5: |
The primary operations on a group are to add and remove members and to discover member objects. These abstract operations, as listed in Figure 6, are implemented in the H5G APIs, as listed in section 4, “Group Function Summaries.”
To add and delete members of a group,
links from the group to existing objects in the file
are created and deleted with the
link and unlink operations.
When a new named object is created,
the HDF5 library executes the link operation in the background
immediately after creating the object
(i.e., a new object is added as a member of the group in which it
is created without further user intervention).
Given the name of an object, the get_object_info method retrieves a description of the object, including the number of references to it. The iterate method iterates through the members of the group, returning the name and type of each object.
| |||||
|
Figure 6: The group object |
Every HDF5 File has a single root group, with the
name /. The root group is identical to any
other HDF5 group, except:
H5Fcreate).
/
reserved for the root group.
New objects are created and then inserted into the graph with a link
operation tht is automatically executed by the library;
existing objects are inserted into the graph with a link operation
explicitly called by the user, which creates a named link from
a group to the object.
An object can be the target of more than one link.
The names on the links must be unique within each group, but there may be many links with the same name in different groups. These are unambiguous, because some ancestor must have a different name, or else they are the same object. The graph is navigated with path names, analogous to Unix file systems (see section 2.3, “HDF5 Path Names”). An object can be opened with a full path starting at the root group, or with a relative path and a starting point. That starting point is always a group, though it may be the current working group, another specified group, or the root group of the file. Note that all paths are relative to a single HDF5 File. In this sense, an HDF5 file is analogous to a single UNIX file system. 1
It is important to note that, just like the UNIX file system, HDF5 objects do not have names, the names are associated with paths. An object has an object identifier that is unique within the file, but a single object may have many names because there may be many paths to the same object. An object can be renamed, or moved to another group, by adding and deleting links. In this case, the object itself never moves. For that matter, membership in a group has no implication for the physical location of the stored object.
Deleting a link to an object does not necessarily delete the object. The object remains available as long as there is at least one link to it. After all links to an object are deleted, it can no longer be opened, although the storage may or may not be reclaimed. 2
It is also important to realize that the linking mechanism can be used to construct very complex graphs of objects. For example, it is possible for object to be shared between several groups and even to have more than one name in the same group. It is also possible for a group to be a member of itself, or to create other cycles in the graph, such as in the case where a child group is linked to one of its ancestors.
HDF5 also has soft links similar to UNIX soft links.
A soft link is an object that has a name and a path name for
the target object. The soft link can be followed to open the target
of the link just like a regular or hard link.
The differences are that the hard link cannot be created if the
target object does not exist and it always points to the same object.
A soft link can be created with any path name, whether or not the
object exists; it may or may not, therefore, be possible to follow
a soft link.
Furthermore, a soft link's target object may be changed.
2.3 HDF5 Path Names
The structure of the HDF5 file constitutes the name space
for the objects in the file.
A path name is a string of components separated by slashes
(/).
Each component is the name of a hard or soft link which points to
an object in the file.
The slash not only separates the components, but indicates their
hierarchical releationship; the component indicated by the link name
following a slash is a always a member of the component indicated by
the link name preceding that slash.
The first component in the path name may be any of the following:
., a period),
indicating the current group
/),
indicating the root group
Component link names may be any string of ASCII characters
not containing a slash or a dot
(/ and ., which are reserved as noted above).
However, users are advised to avoid the use of punctuation and
non-printing characters, as they may create problems for other software.
Figure 7 provides a BNF grammar for HDF5 path names.
PathName ::= AbsolutePathName | RelativePathName
Separator ::= "/" ["/"]*
AbsolutePathName ::= Separator [ RelativePathName ]
RelativePathName ::= Component [ Separator RelativePathName ]*
Component ::= "." | Characters
Characters ::= Character+ - { "." }
Character ::= {c: c Î { { legal ASCII characters } - {'/'} }
|
| Figure 7: A BNF grammar for for HDF5 path names |

|
| Figure 8: An HDF5 file with a directed graph group structure, including a circular reference |
Consider, for example, the file illustrated in Figure 8.
Dataset1 can be identified by either of these
absolute path names:
/GroupA/Dataset1
/GroupA/GroupB/GroupC/Dataset1
Dataset1 can also be identified by this absolute path name:
/GroupA/GroupB/GroupC/GroupA/Dataset1
Alternatively, if the current working location is GroupB,
Dataset1 can be identified by either of these
relative path names:
GroupC/Dataset1
GroupC/GroupA/Dataset1
Note that relative path names in HDF5 do not employ the
../ notation, the UNIX notation indicating a
parent directory, to indicate a parent group.
h5dumph5dump, the command-line utility
distributed with HDF5, to examine a file for purposes either of
determining where to create an object within an HDF5 file or
to verify that you have created an object in the intended place.
inspecting the contents of an HDF5 file.
In the case of the new group created in section 5.1,
“Creating a group,”
the following h5dump command will display the
contents of FileA.h5:
h5dump FileA.h5
Assuming that the discussed objects, GroupA and
GroupB are the only objects that exist in
FileA.h5, the output will look something like the
following:
HDF5 "FileA.h5" {
GROUP "/" {
GROUP GroupA {
GROUP GroupB {
}
}
}
}
h5dump is fully described on the
Tools page of the
HDF5 Reference Manual.
The HDF5 DDL grammar is fully described in the document
DDL in BNF for HDF5,
an element of the HDF5 User's Guide.
|
C Function F90 Function |
Purpose |
H5Gcreate
|
Creates a new empty group and gives it a name. |
H5Gopen
|
Opens an existing group for modification and returns a group identifier for that group. |
H5Gclose
|
Closes the specified group. |
H5Gset_comment
|
Sets the comment for the specified object. |
H5Gget_comment
|
Retrieves the comment for the specified object. |
H5Glink
|
Creates a link of the specified type from a new name to a current name. |
H5Glink2
|
Creates a link of the specified type from a new name to a current name. |
H5Gunlink
|
Removes a link to an object from a group. |
H5Gmove
|
Renames an object within an HDF5 file. |
H5Gmove2
|
Renames an object within an HDF5 file. |
H5Giterate
|
Iterates an operation over the entries of a group. |
(none)
|
Returns the name and type of a specified group member. |
(none)
|
Returns the number of group members. |
H5Gget_objinfo
|
Returns information about an object. |
H5Gget_num_objs
|
Returns number of objects in the specified group. |
H5Gget_objname_by_idx
|
Returns a name of an object specified by its index. |
H5Gget_objtype_by_idx
|
Returns the type of an object specified by its index. |
H5Gget_linkval
|
Returns the name of the object that the specified symbolic link points to. |
The programming model for working with groups is as follows:
To create a group, use H5Gcreate, specifying the
location and the path of the new group.
The location is the identifier of the file or the group in a file
with respect to which the new group is to be identified.
The path is a string that provides wither an absolute path or a
relative path to the new group (see section 2.3,
“HDF5 Path Names”).
A path that begins with a slash (/) is an absolute path
indicating that it locates the new group from the root group of the
HDF5 file.
A path that begins with any other character is a relative path.
When the location is a file, a relative path is a path from that
file's root group;
when the location is a group, a relative path is a path from that group.
The sample code in Figure 9 creates three groups.
The group Data is created in the root directory;
two groups are then created in /Data,
one with absolute path, the other with a relative path.
hid_t file; file = H5Fopen(....); group = H5Gcreate(file, "/Data", 0); group_new1 = H5Gcreate(file, "/Data/Data_new1", 0); group_new2 = H5Gcreate(group, "Data_new2", 0); |
| Figure 9: Creating three new groups |
The third H5Gcreate parameter optionally specifies
how much file space to reserve to store the names that will
appear in this group. If a non-positive value is supplied,
a default size is chosen.
Data.
Any object in a group can be also accessed by its absolute or relative path. To open an object using a relative path, an application must first open the group or file on which that relative path is based. To open an object using an absolute path, the application can use any location identifier in the same file as the target object; the file identifier is commonly used, but object identifier for any object in that file will work. Both of these approaches are illustrated in Figure 10 offers example code in the first two lines to open a group then open a dataset with the appropriate relative path to open the same dataset with an abslolute path and .
Using the file created in the examples above,
Figure 10 provides example code illustrating the use of both
relative and absolute paths to access an HDF5 data object.
The first sequence (two function calls) uses a previously-acquired
file identifier to open the group Data
then uses the returned group identifier and a relative path to open
the dataset CData.
The second approach (one function call) uses the same previously-acquired
file identifier and an absolute path to open the same dataset.
group = H5Gopen(file, "Data"); dataset1 = H5Dopen(group, "CData"); dataset2 = H5Dopen(file, "/Data/CData"); |
| Figure 10: Open a dataset with relative and absolute paths |
/Data.
dataspace = H5Screate_simple(RANK, dims, NULL);
dataset1 = H5Dcreate(file, "/Data/CData", H5T_NATIVE_INT,
dataspace, H5P_DEFAULT);
group = H5Gopen(file, "Data");
dataset2 = H5Dcreate(group, "Cdata2", H5T_NATIVE_INT,
dataspace, plist);
|
| Figure 11: Create a dataset with absolute and relative paths |
H5Gclose ends access to the group
and releases any resources the HDF5 library has maintained
in support of that access, including the group identifier.
As illustrated in Figure 12, all that is required for an
H5Gclose call is the group identifier acquired when
the group was opened; there are no relative versus absolute path
considerations.
herr_t status; status = H5Gclose(group); |
| Figure 12: Close a group |
A non-negative return value indicates that the group was successuflly closed and the resources released; a negative return value indicates that the attempt to close the group or release resources failed.
H5Glink or H5Glink2.
A link is, in effect, is a path
by which the target object can be accessed; it therefore has a
name which functions as a single path component.
A link can be removed with an H5Gunlink call, effectively
removing the target object from the group that contained the link
(assuming, of course, that the removed link was the only link to the
target object in the group).
Hard links
There are two kinds of links, hard links
and soft links.
Hard links are reference counted; soft links are not.
When an object is created, a hard link is automatically created.
An object can be deleted from the file by removing all the hard links to it.
Working with the file from the previous examples,
the code in Figure 13 illustrates the creation of a hard link,
named Data_link, in the root group, /,
to the group Data.
Once that link is created, the dataset Cdata can be accessed
via either of two absolute paths, /Data/Cdata or
/Data_Link/Cdata.
status = H5Glink(file, H5G_LINK_HARD, "Data", "Data_link");
dataset1 = H5Dopen(file, "/Data_link/CData");
dataset2 = H5Dopen(file, "/Data/CData");
|
| Figure 13 |
This and subsequent examples could also use H5Glink2,
which is used exactly like H5Glink except that a
second location identifier is specified and the new object name
is specified relative to the second location identifier.
Figure 14 shows example code to delete a link,
deleting the hard link Data from the root group.
The group /Data and its members are still in the file,
but they can no longer be accessed via a path using the component
/Data.
status = H5Gunlink(file, "Data");
dataset1 = H5Dopen(file, "/Data_link/CData");
/* This call should succeed; all path component still exist*/
dataset2 = H5Dopen(file, "/Data/CData");
/* This call will fail; the path component '/Data' has been deleted*/
|
| Figure 14 |
When the last hard link to an object is deleted, the object is no longer
accessible (although space in the file may not be deallocated).
Figure 15 shows deletion of the last link, Data_link,
to the group originally called Data.
After the unlinking operation, the group is no longer accessible;
consequently, the dataset Cdata is inaccessible.
status = H5Gunlink(file, "Data_link");
dataset = H5Dopen(file, "/Data_link/CData");
/* This call will fail; the dataset is no longer accessible */
|
| Figure 15 |
Soft links
Soft links are objects that assign a name
in a group to a path.
Notably, the target object is determined only when the soft link is accessed,
and may, in fact, not exist. Soft links are not reference counted,
so there may be one or more soft links to an object.
Like hard links, soft links are also created and deleted with
the H5Glink, H5Glink2, and
H5Gunlink functions,
except that soft links are created as type H5G_LINK_SOFT
while hard links are created as type H5G_LINK_HARD.
Returning to our sample file as it was initially created,
Figure 16 shows examples of creating two soft links to the
group /Data.
status = H5Glink(file, H5G_LINK_SOFT, "Data", "Soft2");
status = H5Glink(file, H5G_LINK_SOFT, "Soft2", "Soft3");
dataset = H5Dopen(file, "/Soft2/CData");
|
| Figure 16 |
With the soft links defined in Figure 16, the dataset CData
in the group /Data can now be opened with any of the names
/Data/CData, /Soft2/CData, or
/Soft3/CData.
Note regarding hard links versus soft links
Note that an object's existence in a file is governed by the presence
of at least one hard link to that object.
If the last hard link to an object is removed, the object is removed
from the file and any remaining soft link becomes a
dangling link, a link whose target
object does not exist.
Moving or renaming objects, and a warning
An object can be renamed by changing the name of a link to it with
either H5Gmove or H5Gmove2.
This has the same effect as creating a new link with the new name and
deleting the link with the old name.
Exercise caution in the use of H5Gmove, H5Gmove2
and H5Gunlink as these functions each include a step that
unlinks a pointer to a dataset or group.
If the link that is removed is on the only path leading to an HDF5 object,
that object will become permanently inaccessible in the file.
Consider the following example: assume that the group group2
can only be accessed via the following path, where top_group
is a member of the file‘s root group:
/top_group/group1/group2/
Using H5Gmove or H5Gmove2, top_group
is renamed to be a member of group2. At this point,
since top_group was the only route from the root group to
group1, there is no longer a path by which one can access
group1, group2, or any member datasets.
And since top_group is now a member of group2,
top_group itself and any member datasets have thereby also
become inaccessible.
H5Gget_objinfo function fills this niche by returning a
description of the specified object in an H5G_stat_t structure.
The structure contains the following information:
H5G_GROUP, H5G_DATASET,
H5G_TYPE, or H5G_LINK, respectively
The H5G_stat_t structure specification and the
H5Gget_objinfo function signature appear in Figure 17.
The H5G_stat_t structure elements are as listed above.
The H5Gget_objinfo function parameters are used follows:
loc_id specifies the object for which
information being sought.
name.
follow_link is a Boolean value specifying
whether to follow a soft link and open the target object
(TRUE) or not (FALSE).
H5G_stat_t struct is returned in the
statbuf buffer.
typedef struct H5G_stat_t {
unsigned long fileno[2];
unsigned long objno[2];
unsigned nlink;
int type;
time_t mtime;
size_t linklen;
} H5G_stat_t
herr_t H5Gget_objinfo(hid_t loc_id, const char *name, hbool_t follow_link, H5G_stat_t *statbuf )
|
Figure 17:
The H5G_stat_t struct specification and
the H5Gget_objinfo function signature
|
Figure 18 provides a code example that prints the local paths to the members of a group, following a soft link when it is found.
|
| Figure 18: Printing a specified object's name and type and, in the case of a link, opening the target object |
H5Giterate, is discussed below
H5Giterate is useful both with a single group and
in an iterative process that examines an entire file
or section of a file (the contents of a group, the contents
of all the groups that are members of that group, etc.)
and acts on objects as they are encountered.
An alternative approach is to determine the number of objects
in a group then approach them one at a time.
This is accomplished with the functions
H5Gget_num_objs,
H5Gget_objname_by_idx, and
H5Gget_objtype_by_idx.
H5Gget_num_objs retrieves the number of objects,
say n, in the group.
The values from 0 through n - 1
can then be used as indices to access the members of the group.
For example,
an index value of 0 identifies the first member,
an index value of 1 identifies the second member, and
an index value of n - 1 identifies the last member.
(Note that HDF5 objects do not have permanent indices;
these values are strictly transient and may be different each time a
group is opened.)
Using the index described above, the name and object type can
be retrieved using H5Gget_objname_by_idx and
H5Gget_objtype_by_idx, respectively.
With the name and object type, an application can proceed to
operate as necessary on all or selected group members.
The function H5Giterate, used to discover the members of
a group, is the key to the discovery process.
An application calls H5Giterate with a pointer to a
callback function (see Figure 19).
The HDF5 library iterates through the group specified by the
loc_id and name parameters,
calling the callback function once for each group member.
The callback function must have the signature defined by
H5G_iterate_t.
When invoked, the arguments to the callback function are
the group being iterated,
the group member’s name (the object name), and
a pointer set by the user program.
The callback function is part of the application,
so it can execute any actions the program requires
to discover and store information about the objects.
typedef herr_t (*H5G_iterate_t)(hid_t group_id, const char *member_name,
void *operator_data);
H5Giterate(hid_t loc_id, const char * name, int *idx, H5G_iterate_t operator,
void *operator_data );
|
| Figure 19 |
Note that the H5Giterate function is following the links
from a single group;
these links correspond to the components in a path name.
To iterate over an entire substructure, H5Giterate must be
recursively on every member of the original group that turns out to also
be a group.
To iterate over an entire file, the first call to H5Giterate
must iterate over the root group; subsequent calls to H5Giterate
must then iterate over every subsequent group.
Figure 20 illustrates the relationship between the calling module
of the application, the callback function (do_obj),
and calls to the HDF5 library.
In this diagram, “Global Variables and Functions”
symbolizes the fact that the callback function executes as part
of the application, and may therefore call functions and
update data structures to describe the file and its objects.
|
|
| Figure 20: Relationships between a calling module, the callback function, and the callback function's calls back to the HDF5 library |
Figure 21 illustrates the sequence of events precipitated by an
H5Giterate call.
H5Giterate,
passing a pointer to a callback function
(do_obj in the figure).
Note that the callback function is part of the application.
H5Giterate
call returns to the calling application.
|
|
| Figure 21 |
Figure 22 shows the sequence of calls involved in one iteration of a
callback function that employs the HDF5 function H5Gget_objinfo
to discover properties of the object that is the subject of the current step
of the iteration (e.g., the object’s type and reference count).
The HDF5 library then calls the application’s callback function
do_obj(), which in turn calls the HDF5 library to get the
object information.
The callback function can process the information as needed,
accessing any function or data structure of the application program,
and it can call the HDF5 library again to, for example,
iterate through a group member that is itself a group.
|
|
| Figure 22 |
Over the course of a successful H5Giterate call,
the HDF5 library will call the application’s callback function
once for each member of the group, as illustrated in Figure 23.
At each iteration, the callback function must return a status which
implies a subsequent course of action:
1
Continue iterating.
0
Stop iterating and return to the caller.
Once the iteration has been completed, H5Giterate returns
to the calling application.
|
|
| Figure 23 |
The overall sequence of calls can become quite complex, especially when the callback function in turn calls the HDF5 library. Figure 24 provides a sequence diagram for a case similar to the simple case described above:
H5Giterate on a group,
do_obj once for each group members
(three group members in this case).
H5Gget_objinfo each time it is invoked to discover
information about each object.
|
|
| Figure 24 |
Recursively iterating through the members of every group will result in visiting an object once for each link to it. This may result in visiting an object more than once. The calling application must be prepared to recognize this case and handle it appropriately. If an action should be undertaken only once per object, the application must make sure that it does not repeat the action for an object with two links. For example, if the objects are being copied, it is important that an object with two names be copied once, not twice. Figure 25 illustrates this case.
|
a) The required action is to copy all the objects from one file to another. |
|
|
|
|
| b) A shared dataset should not be copied twice. | c) A shared dataset should be copied once and the apppropriate link should be created. |
| Figure 25 | |
|
|
| Figure 26 |
In such a case, the callback function should check the reference count in
the H5G_stat_t buffer as returned by H5Gget_objinfo.
If the count is greater than one, there is more than one path to
the object in question and it may be in a loop;
the program should act accordingly.
For example, it may be necessary to construct a global table of all the
objects visited.
Note that the object’s name is not unique, but the full path and the
object number (found in the above-mentioned H5G_stat_t buffer)
are unique within an individual HDF5 file.
|
|
|
|
a) The file contains three groups:
the root group, /group1, and /group2.
|
b) The dataset dset1 (or /group1/dset1)
is created in /group1.
|
|
|
|
|
|
c) A link named dset2 to the same dataset
is created in /group2.
|
d) The link from /group1 to dset1 is removed.
The dataset is still in the file, but can be accessed only as
/group2/dset2.
|
|
| Figure 27 | ||
Figure 27 shows examples of the structure of a file with three groups
and one dataset.
The file in Figure 27a contains three groups:
the root group and two member groups.
In Figure 27b, the dataset dset1 has been created
in /group1.
In Figure 27c, a link named dset2 from /group2
to the dataset has been added.
Note that there is only one copy of the dataset;
there are two links to it and it can be accessed either as
/group1/dset1 or as /group2/dset2.
Figure 27d illustrates that one of the two links to the dataset
can be deleted.
In this case, the link from /group1 has been removed.
The dataset itself has not been deleted; it is still in the file
but can only be accessed as /group1/dset2.
|
|
|
|
a) dset1 has two names:
/group2/dset1 and /group1/GXX/dset1.
|
b) dset1 again has two names:
/group1/dset1 and /group1/dset2.
|
|
|
|
|
|
c) dset1 has three names:
/group1/dset1, /group2/dset2,
and /group1/GXX/dset2.
|
d) dset1 has an infinite number of available path names.
|
|
| Figure 28 | ||
Figure 28 illustrates loops in an HDF5 file structure.
The file in Figure 28a contains three groups and a dataset;
group2 is a member of the root group and of the
root group’s other member group, group1.
group2 thus can be accessed by either of two paths:
/group2 or /group1/GXX.
Similarly, the dataset can be accessed either as
/group2/dset1 or as /group1/GXX/dset1.
Figure 28b illustrates a different case: the dataset is a
member of a single group but with two links, or names, in that group.
In this case, the dataset again has two names,
/group1/dset1 and /group1/dset2.
In Figure 28c, the dataset dset1 is a member of two groups,
one of which can be accessed by either of two names.
The dataset thus has three path names: /group1/dset1,
/group2/dset2, and /group1/GXX/dset2.
And in Figure 28d, two of the groups are members of
each other and the dataset is a member of both groups.
In this case, there are an infinite number of paths to the dataset
because GXX and GYY can be traversed
any number of times on the way from the root group, /,
to the dataset.
This can yield a path name such as
/group1/GXX/GYY/GXX/GYY/GXX/dset2.
|
|
|
|
| a) The file contains only hard links. |
b) A soft link is added from group2
to /group1/dset1.
|
|
|
|
|
|
c) A soft link named dset3 is added with a target
that does not yet exist.
| d) Tht target of soft link is created or linked. | |
| Figure 29 | ||
Figure 29 takes us into the realm of soft links.
The original file, in Figure 29a, contains only three hard links.
In Figure 29b, a soft link named dset2 from
group2 to /group1/dset1 has been created,
making this dataset accessible as /group2/dset2.
In Figure 29c, another soft link has been created in group2.
But this time the soft link, dset3, points to a target object
that does not yet exist.
That target object, dset, has been added in Figure 29d
and is now accessible as either /group2/dset or
/group2/dset3.
1It could be said that HDF5 extends the organizing concepts of a file system to the internal structure of a single file.
2As of HDF5-1.4, the storage used for an object is reclaimed, even if all links are deleted.